values must be of shape of (2,1). Got shape(1,2)
软件3.9在做贝叶斯网络values=[[0.6,0.4]]
from pgmpy.models import BayesianModel
from pgmpy.factors.discrete import TabularCPD
from pgmpy.inference import VariableElimination
# 通过边来定义贝叶斯模型
model = BayesianModel([('D', 'G'), ('I', 'G'), ('G', 'L'), ('I', 'S')])
# 定义条件概率分布
cpd_d = TabularCPD(variable='D', variable_card=2, values=[[0.6, 0.4]])
cpd_i = TabularCPD(variable='I', variable_card=2, values=[[0.7, 0.3]])
# variable:变量
# variable_card:基数
# values:变量值
# evidence:
cpd_g = TabularCPD(variable='G', variable_card=3,
values=[[0.3, 0.05, 0.9, 0.5],
[0.4, 0.25, 0.08, 0.3],
[0.3, 0.7, 0.02, 0.2]],
evidence=['I', 'D'],
evidence_card=[2, 2])
cpd_l = TabularCPD(variable='L', variable_card=2,
values=[[0.1, 0.4, 0.99],
[0.9, 0.6, 0.01]],
evidence=['G'],
evidence_card=[3])
cpd_s = TabularCPD(variable='S', variable_card=2,
values=[[0.95, 0.2],
[0.05, 0.8]],
evidence=['I'],
evidence_card=[2])
# 将有向无环图与条件概率分布表关联
model.add_cpds(cpd_d, cpd_i, cpd_g, cpd_l, cpd_s)
# 验证模型:检查网络结构和CPD,并验证CPD是否正确定义和总和为1
model.check_model()
# 获得G点的概率表
print(model.get_cpds('G'))
# 获得G点的基数
print(model.get_cardinality('G'))
# 获取贝叶斯网络的局部依赖关系
print(model.local_independencies(['D','I','S','G','L']))
# 贝叶斯推理:变量消除
infer = VariableElimination(model)
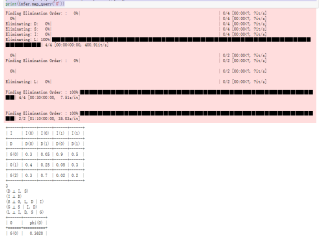
print(infer.query(['G']), ['G'])
print(infer.query(['G'], evidence={'D':0, 'I':1})['G'])
print(infer.map_query('G'))
from pgmpy.models import BayesianModel
from pgmpy.factors.discrete import TabularCPD
from pgmpy.inference import VariableElimination
# 通过边来定义贝叶斯模型
model = BayesianModel([('D', 'G'), ('I', 'G'), ('G', 'L'), ('I', 'S')])
# 定义条件概率分布
cpd_d = TabularCPD(variable='D', variable_card=2, values=[[0.6], [0.4]])
cpd_i = TabularCPD(variable='I', variable_card=2, values=[[0.7], [0.3]])
# variable:变量
# variable_card:基数
# values:变量值
# evidence:
cpd_g = TabularCPD(variable='G', variable_card=3,
values=[[0.3, 0.05, 0.9, 0.5],
[0.4, 0.25, 0.08, 0.3],
[0.3, 0.7, 0.02, 0.2]],
evidence=['I', 'D'],
evidence_card=[2, 2])
cpd_l = TabularCPD(variable='L', variable_card=2,
values=[[0.1, 0.4, 0.99],
[0.9, 0.6, 0.01]],
evidence=['G'],
evidence_card=[3])
cpd_s = TabularCPD(variable='S', variable_card=2,
values=[[0.95, 0.2],
[0.05, 0.8]],
evidence=['I'],
evidence_card=[2])
# 将有向无环图与条件概率分布表关联
model.add_cpds(cpd_d, cpd_i, cpd_g, cpd_l, cpd_s)
# 验证模型:检查网络结构和CPD,并验证CPD是否正确定义和总和为1
model.check_model()
# 获得G点的概率表
print(model.get_cpds('G'))
# 获得G点的基数
print(model.get_cardinality('G'))
# 获取贝叶斯网络的局部依赖关系
print(model.local_independencies(['D','I','S','G','L']))
# 贝叶斯推理:变量消除
infer = VariableElimination(model)
print(infer.query(['G']), ['G'])
print(infer.query(['G'], evidence={'D':0, 'I':1})['G'])
print(infer.map_query('G'))cpd_d = TabularCPD(variable='D', variable_card=2, values=[[0.6], [0.4]])
cpd_i = TabularCPD(variable='I', variable_card=2, values=[[0.7], [0.3]])
他要一维数组,你给了二维数组。
你的类型传递错误了。 建议将代码贴全。
shape of (2,1). Got shape(1,2) 肯定是数据类型写反了。
您好,我是有问必答小助手,您的问题已经有小伙伴解答了,您看下是否解决,可以追评进行沟通哦~
如果有您比较满意的答案 / 帮您提供解决思路的答案,可以点击【采纳】按钮,给回答的小伙伴一些鼓励哦~~
ps:问答VIP仅需29元,即可享受5次/月 有问必答服务,了解详情>>>https://vip.csdn.net/askvip?utm_source=1146287632