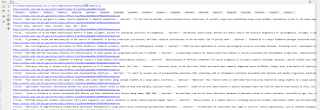
用于抓取文章的代码,爬取时报错,请解决
import requests
from bs4 import BeautifulSoup
def search_pubmed(query):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
search_url = base_url + "esearch.fcgi?db=pubmed&term=" + query
response = requests.get(search_url)
soup = BeautifulSoup(response.text, '')
id_list = [id.text for id in soup.find_all('Id')]
return id_list
def fetch_details(pubmed_id):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
fetch_url = base_url + "efetch.fcgi?db=pubmed&id=" + pubmed_id + "&retmode=xml"
response = requests.get(fetch_url)
soup = BeautifulSoup(r.text, 'html.parser')
soup = BeautifulSoup(response.text, 'xml')
try:
title = soup.find('ArticleTitle').text
except AttributeError:
title = None
try:
abstract = soup.find('AbstractText').text
except AttributeError:
abstract = None
try:
journal = soup.find('JournalTitle').text
except AttributeError:
journal = None
try:
doi = soup.find('ArticleId', {'IdType': 'doi'}).text
except AttributeError:
doi = None
return {'title': title, 'abstract': abstract, 'journal': journal, 'doi': doi}
# Example usage
ids = search_pubmed('human')
for id in ids:
details = fetch_details(id)
print(details)
报错如下;
Traceback (most recent call last):
File "F:/桌面/抓2.py", line 38, in <module>
ids = search_pubmed('human')
File "F:/桌面/抓2.py", line 9, in search_pubmed
soup = BeautifulSoup(response.text, '')
File "C:\Users\HUAWEI\AppData\Local\Programs\Python\Python311\Lib\site-packages\bs4\__init__.py", line 249, in __init__
raise FeatureNotFound(
bs4.FeatureNotFound: Couldn't find a tree builder with the features you requested: . Do you need to install a parser library?
已经安装1xlm仍然报错,求解
代码 存在问题,已经帮你修改好了,下面的代码能够正常运行。
如果对你有帮助,点个采纳谢谢!
# !/usr/bin/env python
# -*- coding: utf-8 -*-
# @author: yjp
# @software: PyCharm
# @file: main.py
# @time: 2022-08-08 16:49
import requests
from bs4 import BeautifulSoup
def search_pubmed(query):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
search_url = base_url + "esearch.fcgi?db=pubmed&term=" + query
print(search_url)
response = requests.get(search_url)
soup = BeautifulSoup(response.text, 'xml')
id_list = [id.text for id in soup.find_all('Id')]
print(id_list)
return id_list
def fetch_details(pubmed_id):
base_url = "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/"
fetch_url = base_url + "efetch.fcgi?db=pubmed&id=" + pubmed_id + "&retmode=xml"
print(fetch_url)
response = requests.get(fetch_url)
soup = BeautifulSoup(response.text, 'xml')
try:
title = soup.find('ArticleTitle').text
except AttributeError:
title = None
try:
abstract = soup.find('AbstractText').text
except AttributeError:
abstract = None
try:
journal = soup.find('JournalTitle').text
except AttributeError:
journal = None
try:
doi = soup.find('ArticleId', {'IdType': 'doi'}).text
except AttributeError:
doi = None
return {'title': title, 'abstract': abstract, 'journal': journal, 'doi': doi}
if __name__ == '__main__':
# Example usage
ids = search_pubmed('human')
for id in ids:
details = fetch_details(id)
print(details)